Interview | AI Research Insights
.png)
As Visage’s Clinical Research Director North America [@MingDeLin1234], I have the privilege to work with numerous clinical and scientific leaders across the Visage 7 customer base. The scope of research initiatives is exhilarating, and the possibilities are in many ways, endless. Of course, it’s thrilling to be able to report on new features, algorithms and applications that may be introduced into clinical production, that’s everyone’s ultimate end goal. However, the research journey has twists and turns along the way, often with the creation of new capabilities, some incremental and some maybe a bit bigger. Sometimes those capabilities are spawned out of inspiration and sometimes out of necessity, but importantly they represent advancements that have a positive impact on other research initiatives with similar needs. This collectively has the ability to move multiple Visage customer research initiatives forward using the Visage AI Accelerator program.
A rising research star is Mariam Aboian, MD, PhD, Assistant Professor, Radiology & Biomedical Imaging, Yale School of Medicine. I had the recent opportunity to interview Dr. Aboian [@MariamAboian] about her experiences and research journey working with Visage. As I did, I hope you enjoy her comprehensive insights which may prove useful to your own research initiatives. If you have any comments or questions, drop me an email so we can continue the conversation.
Q: Thank you for joining us on the Visage Blog. What is your research project about, what is its clinical relevance, and clinical challenges this research is aiming to overcome? Can you also discuss the end goal(s) of the project (e.g. embedding algorithm into diagnostic workflow for routine clinical use)?
The goal of our research project is to incorporate quantitative imaging tools into clinical practice in neuroradiology. The project originated from a very simple problem that we were struggling in implementing in pediatric neuroradiology. We needed a method for reliable and fast volume measurements for pediatric brain tumors. This was not available in clinical practice because some of the viewers would not measure volume and in others, the measurement would just take too long. Due to these limitations, very complex and heterogeneous tumors were compared using 2D measurements and visual assessment. When I started at Yale, I was using the Viewer Objects (Overlay) tool in Visage 7 that allows comparison of tumor volumes. This tool was a major game changer in my practice and I currently use it for all my brain tumor follow up studies and also for hydrocephalus studies. This tool was ideal, but I still needed to compare volumes of multiple prior imaging studies and develop growth curves for follow up of tumor size during treatment.
At that point, we reached out to Visage with the goal to develop an artificial intelligence (AI) based auto-segmentation tool for brain tumors that is embedded into the Visage 7 Research Server. With your help, as well as help from Khaled Bousabarah, PhD and Wolfgang Holler, we now have a new GliomaSegmentation tool available for auto-segmentation of high-grade gliomas. The tool works as a button that is plugged into the software and would auto-segment the tumor on FLAIR and T1 post-gadolinium sequences, generating 3D segmentation of Whole Tumor, Core (contrast enhancing portion of the tumor), and Necrotic (the central necrosis within the tumor).
Q: How was your research on a technical/tooling level being conducted and some of the frustrations/hurdles you experienced prior to working with Visage? For example, how many different software(s) did you use? Were they homegrown, open source or commercial? What was manual (vs. semi-/fully-automatic)? What aspects of the tech/tooling were most onerous in terms of the project efficiency and effort pre-Visage?
One of the major limitations to doing quantitative neuroimaging is the availability of tools for image processing and understanding of how to develop image processing tools. I do not have a computer science background, so development of new homegrown image processing tools in my laboratory has been difficult for me. I tried a number of methods in the past – Horos freeware, 3D slicer, as well as an advanced visualization viewer from a major modality vendor. I also had Computer Science (CS) friends code image processing tools for me in MATLAB. I would encounter major limitations in terms of difficulty transferring images from PACS to the software, de-identification of images for DICOM transfer, lack of intuitive steps for image processing, running into dead-ends and trying to figure out workarounds to solve the problems using the Internet, the time it took to segment the tumors, and the ability to extract quantitative information for image analysis. It always seemed that I needed a laboratory that could perform these tasks for me, but the groups that could develop these tools were either at different institutions and had their own ‘to do lists’, or were very busy, and had to pursue their own research projects.
I had very important projects that needed quantitative imaging tools, but I had no way to answer the questions until these tools were available. This is the reason why I am very grateful to the Visage product development team, who both listen to my needs as a researcher and have worked to develop the tools that we need in clinical practice. For example, I remember when we started the project we were trying to develop an ADC histogram extraction method from diffusion weighted imaging. I reached out to Visage, explained the workflow and information we would need to extract. The tool was ready within a few weeks and we were able to extract ADC histograms running the code on Jupyter Notebook. Furthermore, it was important for us to not export the image processing outside of clinical workflow, so we developed plugins for feature extraction.
In addition to the auto-segmentation button, we also have a PyRadiomics extraction button within the Visage 7 Research Server, that automatically extracts imaging features from the segmentation into a JSON file. We are also exploring methods how to extract features with a consistent naming process, so that when we get our NIfTI files extracted, we know which segmentation it is and on which sequence.
This brings us to the copy/paste tool (e.g., Quad MPR comparison layout) that was developed by the Visage team for us. This tool is very exciting because when I segment the tumor on one sequence, I need to use the same tumor segmentation in a different sequence. If I re-draw the segmentation, it is not identical. If I copy/paste the segmentation, it is commonly misregistered and requires a lot of time for fixing the segmentation. The new tool that was developed automatically aligns all of the sequences and then the segmented ROI is copied and pasted on other sequences within the same layout. With this feature, I no longer have to go in and fix the segmentations, and I am also able to extract features from multiple sequences using the same ROI. The copy/paste of my segmentation now takes seconds.
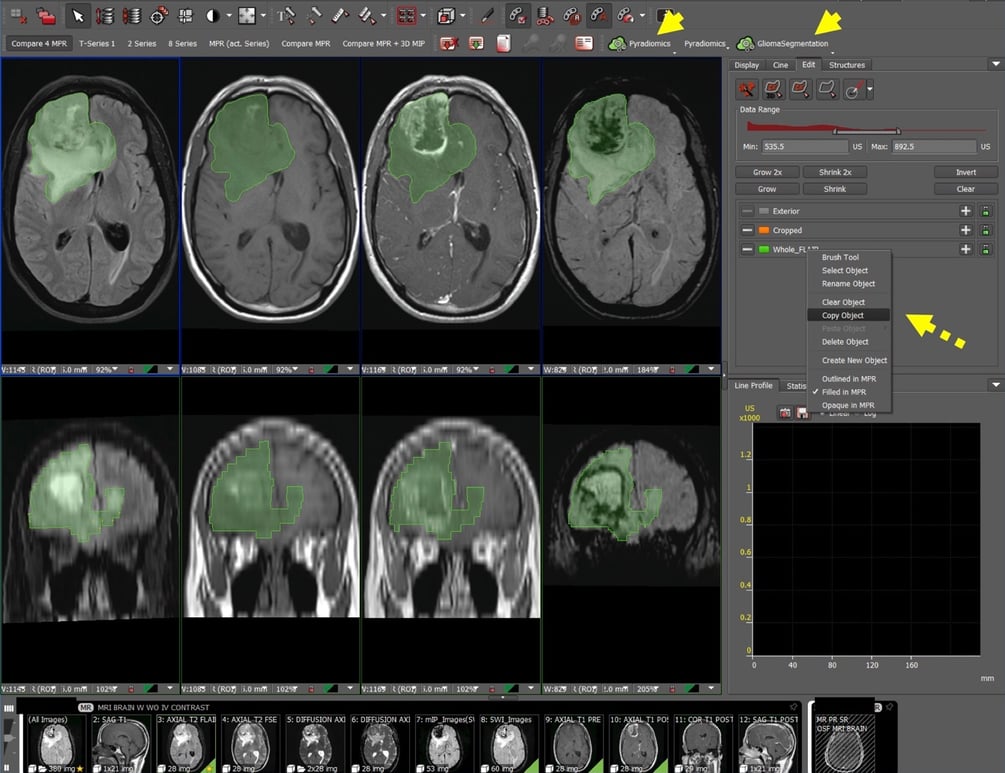
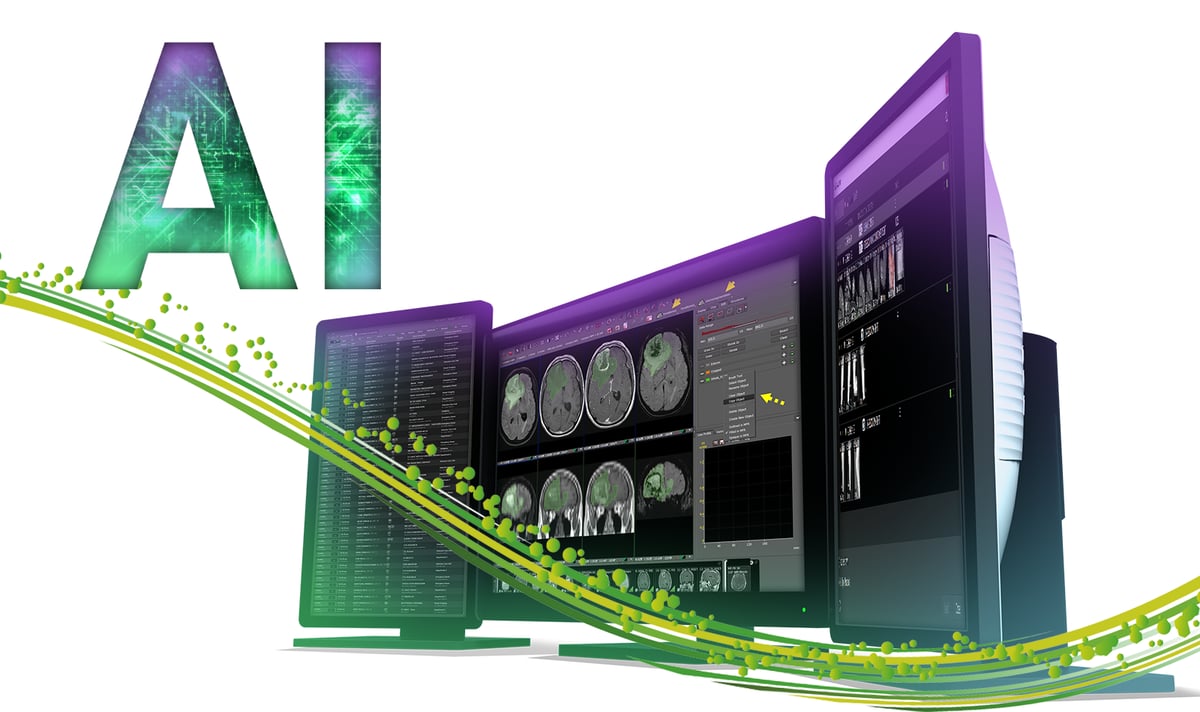
Figure 1: Auto-segmentation and feature extraction tool on Visage 7 Research Server allows incorporation of radiomic-based analysis into clinical workflow. Auto-segmentation and PyRadiomics feature extraction buttons are incorporated into the user interface (yellow arrowheads). The auto-segmentation of tumor and surrounding edema is performed on FLAIR images and can be copied onto other sequences using the Copy Object tool (yellow dashed arrow).
Q: How have the new features/capabilities been helpful in accelerating/streamlining the research process? For example, Quad MPR comparison layout, native PyRadiomics embedding, AI-based brain tumor segmentation, export of segmentation masks as NIfTIs, etc.
One of the very first tools we are developing for use at Yale is a method to predict high- versus low-grade glioma. We were able to segment 120 high-grade gliomas using the auto segmentation tool and extract imaging features from T1, T2, FLAIR, T1PG spin echo, T1PG gradient echo, ADC, and SWI sequences that will be used in machine learning based analysis. We are currently confirming the segmentations of a low-grade glioma cohort for that analysis. For the high-grade glioma dataset, we were able to batch extract the NIfTI files and upload them into our research folder for training our algorithm. We also used the native PyRadiomics button in Visage to extract radiomic features from the segmentations. We confirmed that extracting PyRadiomics features from the Visage 7 Research Server is the same as extracting the features from the NIfTI file of the segmentation exported from Visage. In the future, we plan to only use the built-in PyRadiomics button for feature extraction. Currently, we are further optimizing the use of the PyRadiomics button.
Q: Can you please provide an overview of how to do glioma measurements in Visage with figures and captions?
To measure high-grade glioma Whole, Core, and Necrotic portions, we first open the study in Visage and press the GliomaSegmentation button. We then open up the Quad MPR (Compare 4 MPR) layout to automatically align the images. The segmentation can then be copied and pasted onto the other sequences using the copy/paste Copy Object tool. We also check the accuracy of the paste and modify as needed. After that is set up, we can either click the PyRadiomics button to download the JSON file from the 3D segmentations, or extract the NIfTI file for further offline processing.
Q: In addition to the technical advancements, can you describe your most memorable experiences working with the Visage development team?
I am very grateful for the receptiveness and support that I get from the Visage team. I feel that I am being listened to and encouraged to succeed. I see the Visage team as an essential partner for my laboratory and for the development of imaging tools that we want to translate into clinical practice.
[Note: The Visage AI Accelerator program is an end-to-end AI solution that bridges research and diagnostic imaging on the same, unified platform. Some of the capabilities described above, including PyRadiomics and GliomaSegmentation, are intended for use with the Visage 7 Research Server.]